Topic 2 Data Input and Output in R
Loading data into R can be quite frustrating since there so many different ways available for different types data files. We compile a short note to list some of these functions that commonly used to read the most commonly used formats of data in practice. We may also want to use some data sets built in some libraries (packages) in R for the purpose of illustration and practice.
2.1 Built-in Data
Build-in data are also ready in R format. To get the list of available data sets in base R, we can use data(). However, if we want to get the list of data sets available in a package, we first need to load that package then data() command shows the available data sets in that package.
For example, the library datasets in base R has a number of built-in data sets, we can use command data() or ls("package:datasets") to list all of these built-in data sets. When opening the R session, all built-in data sets in library datasets are all automatically loaded to the current working directory. We can simply use any of these data set The well-known iris is one of these built-in data sets. Next, we make a pair-wise scatter plot for all numerical variables in the iris data set in the following
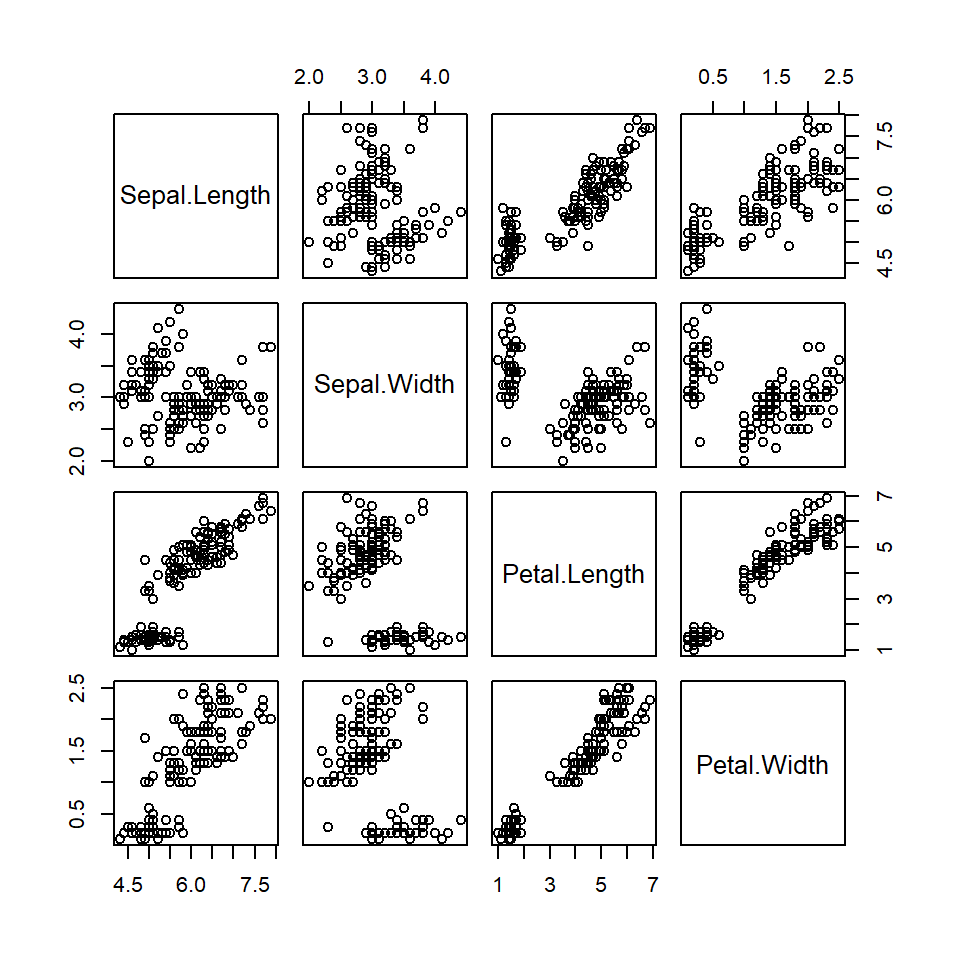
Figure 2.1: Pair-wise scatter plot of iris data
To use a built-in data set that is not in the library datasets in base R, we need to load the library first. After the library is loaded, we can use command to list all data sets in that library. For example, PimaIndiansDiabetes2 is a frequently used data set in logistic regression modeling. It is a built-in data set in the library mlbench. If we don’t load library mlbench (Machine Learning Benchmark Problems), we will not be able to access that data set.
summary(PimaIndiansDiabetes2)The message returned from the above code is Error in summary(PimaIndiansDiabetes2): object 'PimaIndiansDiabetes2' not found. However, if we load the library first, then generate the summary table, we will have
library(knitr)
if (!require("mlbench")) { # check whether *mlbench* is installed in the machine
install.packages("mlbench") # if not, install it
library(mlbench) # then load the package!
}
data(PimaIndiansDiabetes2) # load the specific data set to the working directory
summary(PimaIndiansDiabetes2) # use the data set for analysis## pregnant glucose pressure triceps
## Min. : 0.000 Min. : 44.0 Min. : 24.00 Min. : 7.00
## 1st Qu.: 1.000 1st Qu.: 99.0 1st Qu.: 64.00 1st Qu.:22.00
## Median : 3.000 Median :117.0 Median : 72.00 Median :29.00
## Mean : 3.845 Mean :121.7 Mean : 72.41 Mean :29.15
## 3rd Qu.: 6.000 3rd Qu.:141.0 3rd Qu.: 80.00 3rd Qu.:36.00
## Max. :17.000 Max. :199.0 Max. :122.00 Max. :99.00
## NA's :5 NA's :35 NA's :227
## insulin mass pedigree age diabetes
## Min. : 14.00 Min. :18.20 Min. :0.0780 Min. :21.00 neg:500
## 1st Qu.: 76.25 1st Qu.:27.50 1st Qu.:0.2437 1st Qu.:24.00 pos:268
## Median :125.00 Median :32.30 Median :0.3725 Median :29.00
## Mean :155.55 Mean :32.46 Mean :0.4719 Mean :33.24
## 3rd Qu.:190.00 3rd Qu.:36.60 3rd Qu.:0.6262 3rd Qu.:41.00
## Max. :846.00 Max. :67.10 Max. :2.4200 Max. :81.00
## NA's :374 NA's :11We can also use data() to list all buit-in data set in the package
library(mlbench) # load the package available in the machine
data(package = 'mlbench') # list all data sets available in the packageNote: if the package name is not specified in the argument of data(), all built-in data sets in all loaded packages will be listed!
2.2 Loading External Data Sets
Loading an external data set to can be a challenge depending on the format and the structure of the data.
Before moving on and discover how to load data into R, it might be useful to go over the following checklist that will make it easier to import the data correctly into R:
If working with spreadsheets, the first row is usually reserved for the header, while the first column is used to identify the sampling unit;
Avoid names, values, or fields with blank spaces. Otherwise, each word will be interpreted as a separate variable, resulting in errors that are related to the number of elements per line in your data set;
If we want to concatenate words, inserting a
.in between two words instead of a space; Short names are preferred over longer names;Try to avoid using names that contain symbols, such as ?, $,%, ^, &, *, (, ),-,#, ?,,,<,>, /, |, , [ ,] ,{, and };
Delete any comments in the Excel file to avoid extra columns or NA’s to be added to the file; and
Make sure that any missing values in the data set are indicated with NA.
2.2.1 Preparing R Workspace
It is a good practice to empty all R objects defined in other R sessions to avoid miss-use (unintentional-use) of R objects with simple easy names that were defined in the other sessions.
We might want to start an environment that is NOT filled with data and values from other sessions. The simplest way is to delete all existing R objects before starting new analysis using the following line of code
The rm() function removes objects from a specified environment. We can then use ls() to check whether all previously defined R objects were removed.
2.2.2 Setting-up Working Directory
It is dependent on whether you are writing R script or RMarkdown. If writing an R script, it is suggested to set-up a working directory for the specific analysis task using the following R function
setwd("<location of your dataset>")If writing RMarkdown document, by the default, the folder we save the RMarkdown document is the automatically set as working directory (also called document directory). setwd() in the code chunk does not work in RMarkdown!
2.3 Read Common Files into R
The following basic R functions focus on getting spreadsheets into R, rather than Excel or other type of files. There are ways of importing other files into R using various R functions in different packages.
2.3.1 Read TXT files with read.table()
If you have a .txt or a tab-delimited text file, we can easily import it with the basic R function read.table().
- Data with No Column Names
In other words, the contents of the file will look similar to this
1 6 a
2 7 b
3 8 c
4 9 d
5 10 eIf the above data file is saved in a local folder, say, C:\peng\eBooks\STA321\fakeDat.txt, we use the following code to load this data to R
myFakeData <- read.table("C:\\peng\\eBooks\\STA321\\fakeDat.txt")If the above data file is on a web server, say, http://www.someserver.com/STA321/fakeDat.txt, we use the following code to load this data to R
myFakeData <- read.table("http://www.someserver.com/STA321/fakeDat.txt")- Data with Good Column Names
The above fake data file does not have column names, the loaded R data set will be automatically assigned names V1, V2, and V3 to the three corresponding columns. If the data file has column names and we want to use it in the R data frame, we need to use the header argument.
ID num char
1 6 a
2 7 b
3 8 c
4 9 d
5 10 eWe can use the following code to load the data correctly in R.
myFakeData <- read.table("C:\\peng\\eBooks\\STA321\\fakeDat.txt", header = TRUE)or
myFakeData <- read.table("http://www.someserver.com/STA321/fakeDat.txt", header = TRUE)- Data With Bad Column Names
Sometimes, we may have a data file with messy or unwanted column names, some comments about the data set, we can use skip argument to skip certain number of rows when read the data file to R then rename the columns with appropriate variable names. For example, if the data file has comments like
This data set was collected by someone in the cloud.
the column names also violates the naming convention.
ID num@3 char#
1 6 a
2 7 b
3 8 c
4 9 d
5 10 eWe can read the above data in R and rename the columns using the following code
myFakeData <- read.table("C:\\peng\\eBooks\\STA321\\fakeDat.txt", skip = 3)
names(myFakeData) = c("var1", "var2", "var3")2.4 Data Values Separated By Special Symbol
If the data file has a special separator, we need to use argument sep= to specify the separator.
This data set was collected by someone in the cloud.
the column names also violates the naming convention.
ID @ num @ char
1 @ 6 @ a
2 @ 7 @ b
3 @ 8 @ c
4 @ 9 @ d
5 @ 10 @ eThe following code will accomplish the task.
myFakeData <- read.table("C:\\peng\\eBooks\\STA321\\fakeDat.txt", header = TRUE, skip = 2, sep = "@")2.4.1 Importing a CSV into R
If the values were separated with a , or ;, we usually are working with a .csv file. Its contents will look similar to this:
Col1,Col2,Col3
1, 2, a
4, 5, b
7, 8, dTo successfully load this file into R, we can also use the read.table() function in which we specify the separator character, or we can use the read.csv() or read.csv2() functions. The former function is used if the separator is a ,, the latter if ; is used to separate the values in your data file.
Remember that the read.csv() as well as the read.csv2() function are almost identical to the read.table() function, with the sole difference that they have the header and fill arguments set as TRUE by default.
2.5 Import SAS, SPSS, and Other Data Sets into R
R is a programming language and software environment for statistical computing. sometimes we need to import data from advanced statistical software programs such as SAS and SPSS. We need to install specialized packages to achieve this task. The well-known package foreign has several R function to read different data sets generated from different software programs.
2.5.1 Import SPSS Files into R
The R package foreign has a function read.spss() to read SPSS data set (with extension .sav). The code is something like
# Activate the `foreign` library
library(foreign)
# Read the SPSS data
mySPSSData <- read.spss("example.sav")If we want the result to be displayed in a data frame, we can set the to.data.frame argument of the read.spss() function to TRUE. Furthermore, if we do NOT want the variables with value labels to be converted into R factors with corresponding levels, we should set the use.value.labels argument to FALSE:
# Activate the `foreign` library
library(foreign)
# Read the SPSS data
mySPSSData <- read.spss("example.sav",
to.data.frame=TRUE,
use.value.labels=FALSE)Remember that factors are variables that can only contain a limited number of different values. As such, they are often called “categorical variables”. The different values of factors can be labeled and are therefore often called “value labels”
2.5.2 mport Stata Files into R
To import Stata files, we use the read.dta() function in package foreign to read data into R:
# Activate the `foreign` library
library(foreign)
# Read Stata data into R
mydata <- read.dta("<Path to file>") 2.5.3 Import Systat Files into R
If you want to get Systat files into R, we also want to use the foreign package, just like shown below:
# Activate the `foreign` library
library(foreign)
# Read Systat data
mydata <- read.systat("<Path to file>") 2.5.4 Import SAS Files into R
We need to install the sas7bdat package and load it, and then invoke the read.sas7bdat() function contained within the package.
# Activate the `sas7bdat` library
library(sas7bdat)
# Read in the SAS data
mySASData <- read.sas7bdat("example.sas7bdat")Note that we can also use the foreign library to load in SAS data in R. In such cases, we’ll start from a SAS Permanent Dataset or a SAS XPORT Format Library with the read.ssd() and read.xport() functions, respectively. But this is relatively inconvenient.